Replies: 3 comments 4 replies
-
|
Hi @mm809, I'm happy to hear that you want to try Cell-ACDC! It's certainly possible to load and correct the result of an external tracker. It's also possible to add external trackers directly into Cell-ACDC. Which tracker did you use? Anyway, to load the result of an external tracker, you will need to convert it to a format compatible with Cell-ACDC. What is the output of this tracker? Do you have the segmentation masks? The segmentation mask for Cell-ACDC is a NumPy file saved as Cheers, |
Beta Was this translation helpful? Give feedback.
-
|
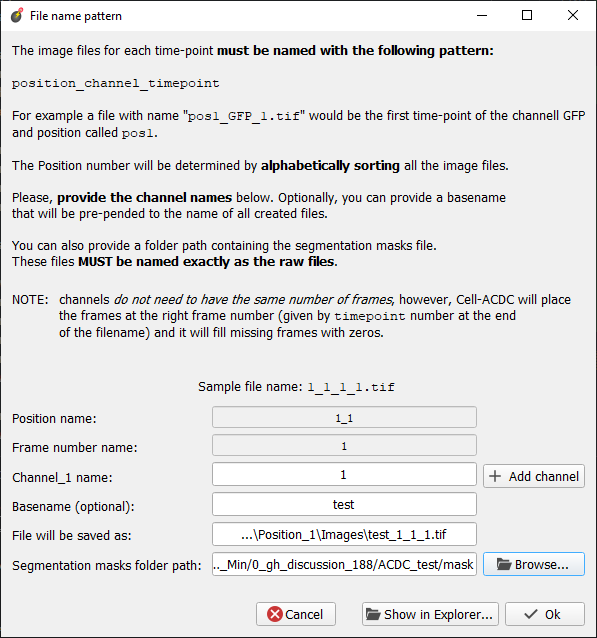
Hi @mm809, Thank you for sending me your data, I released To update to the latest version, please activate the acdc environment and the run the command Next, run module 0. and instead of Bio-Formats choose "Re-structure files". Go through the pop-ups and you should get the Position folders that you can load into Cell-ACDC. I attached a screenshot of the parameters for the conversion that I used with your test data. Let me know if it's clear enough and if it works, thanks! Cheers, |
Beta Was this translation helpful? Give feedback.
-
|
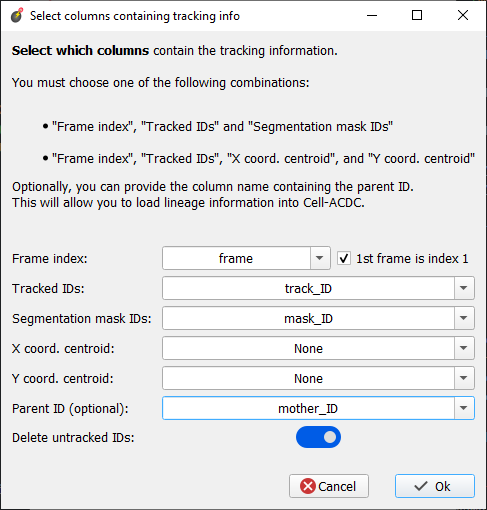
Hi @mm809 I just released You can run the new utility for the menu on the top menubar of the Cell-ACDC launcher called Next, go through the wizard and select a Position folder (one with already the segmentation mask in Cell-ACDC format). Hopefully, the process will be self-explanatory. I attached here a screenshot of the main window where you select which column is what with the selection I used. Let me know if it works or if you have issues, thanks! Cheers, PS: For the correction of the mother-daughter relationships I think we will need to discuss a bit what you want to achieve because I think it will need some fine-tuning. Nobody started from pre-annotated data so far. We can do that in another issue. Feel free to open it when you are ready. |
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
-
Hello,
Thanks for sharing this neat GUI! Would it be possible to use it for correcting tracking errors from an external tracker? It seems that it might be doable if one converts lineage information into the masks and .h5 files in the same format as the segmentation GUI output. Thanks!
Beta Was this translation helpful? Give feedback.
All reactions