Important
We are very sorry that the old link for RNABPDB was not functional for some time. Now, you can find the database again at our new addresses:
or
RNAHelix server is available at :
or
RNA Base Pair Databae raw flat files for version 1 and version 2 is stored as tab seperated format. Please follow references above for parameter description.
Pdb Information tab is representing : pdbid_BasePair_Pairing Edge_1st residue number according to bpfind_2nd residue number according to bpfind_1st residue number according to PDB_1st Chain id according to bpfind_2nd residue number according to PDB_2nd Chain id according to bpfind
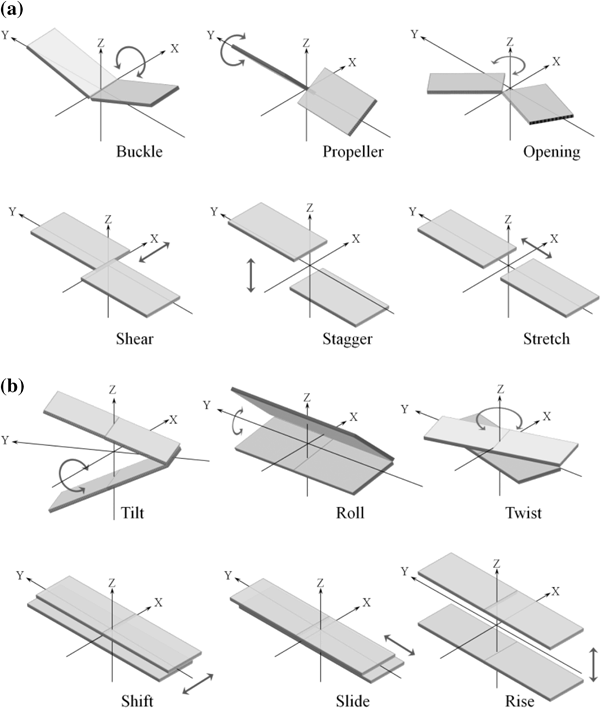
| Buckle | Open | Propeller | Stagger | Shear | Stretch | C1'-C1'_Distance | Evalue | Proximity | Pdb Information |
|---|---|---|---|---|---|---|---|---|---|
| -29.12 | 15.47 | -22.86 | -0.80 | 2.32 | 2.68 | 0.63 | 12.52 | 9.68 | 6ys3_AA_wwC_2919_2993_2817_b_2891_b |
| Tilt | Roll | Twist | Shift | Slide | Rise | Cup | Overlap | Alpha11 | Beta11 | Gamma11 | Chi11 | Eta11 | Theta11 | Phase11 | Eps12 | Zeta12 | Chi12 | Phase12 | C1c111 | Alpha21 | Beta21 | Gamma21 | Chi21 | Eta21 | Theta21 | Phase21 | Eps22 | Zeta22 | Chi22 | Phase22 | C1c121 | Pdb Information |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.65 | 12.52 | 34.84 | 1.29 | -2.84 | 3.03 | -8.41 | 37.18 | -57.20 | 142.70 | 59.60 | -159.90 | 153.80 | -118.00 | 11.80 | -141.80 | -72.90 | -177.20 | 14.50 | 5.89 | -60.90 | 171.50 | 52.10 | -159.40 | 165.10 | -175.80 | 16.40 | -145.80 | -68.20 | -170.10 | 12.00 | 6.15 | 6zm6_AA_wwC_256_308_1926_A_1978_A |